CASCADE 2.0 Analysis (Topology and Link Operator Mutations)
# 'ss' => calibrated models, 'rand' => proliferative models (so not random but kind of!)
# 'ew' => ensemble-wise, 'mw' => modelwise
## HSA results ss
topolink_ss_hsa_ew_50sim_file = paste0("results/topo-and-link/cascade_2.0_ss_50sim_fixpoints_hsa_ensemblewise_synergies.tab")
topolink_ss_hsa_mw_50sim_file = paste0("results/topo-and-link/cascade_2.0_ss_50sim_fixpoints_hsa_modelwise_synergies.tab")
topolink_ss_hsa_ew_150sim_file = paste0("results/topo-and-link/cascade_2.0_ss_150sim_fixpoints_hsa_ensemblewise_synergies.tab")
topolink_ss_hsa_mw_150sim_file = paste0("results/topo-and-link/cascade_2.0_ss_150sim_fixpoints_hsa_modelwise_synergies.tab")
topolink_ss_hsa_ew_synergies_50sim = emba::get_synergy_scores(topolink_ss_hsa_ew_50sim_file)
topolink_ss_hsa_mw_synergies_50sim = emba::get_synergy_scores(topolink_ss_hsa_mw_50sim_file, file_type = "modelwise")
topolink_ss_hsa_ew_synergies_150sim = emba::get_synergy_scores(topolink_ss_hsa_ew_150sim_file)
topolink_ss_hsa_mw_synergies_150sim = emba::get_synergy_scores(topolink_ss_hsa_mw_150sim_file, file_type = "modelwise")
## HSA results rand
topolink_prolif_hsa_ew_50sim_file = paste0("results/topo-and-link/cascade_2.0_rand_50sim_fixpoints_hsa_ensemblewise_synergies.tab")
topolink_prolif_hsa_mw_50sim_file = paste0("results/topo-and-link/cascade_2.0_rand_50sim_fixpoints_hsa_modelwise_synergies.tab")
topolink_prolif_hsa_ew_150sim_file = paste0("results/topo-and-link/cascade_2.0_rand_150sim_fixpoints_hsa_ensemblewise_synergies.tab")
topolink_prolif_hsa_mw_150sim_file = paste0("results/topo-and-link/cascade_2.0_rand_150sim_fixpoints_hsa_modelwise_synergies.tab")
topolink_prolif_hsa_ew_synergies_50sim = emba::get_synergy_scores(topolink_prolif_hsa_ew_50sim_file)
topolink_prolif_hsa_mw_synergies_50sim = emba::get_synergy_scores(topolink_prolif_hsa_mw_50sim_file, file_type = "modelwise")
topolink_prolif_hsa_ew_synergies_150sim = emba::get_synergy_scores(topolink_prolif_hsa_ew_150sim_file)
topolink_prolif_hsa_mw_synergies_150sim = emba::get_synergy_scores(topolink_prolif_hsa_mw_150sim_file, file_type = "modelwise")
## Bliss results ss
topolink_ss_bliss_ew_50sim_file = paste0("results/topo-and-link/cascade_2.0_ss_50sim_fixpoints_bliss_ensemblewise_synergies.tab")
topolink_ss_bliss_mw_50sim_file = paste0("results/topo-and-link/cascade_2.0_ss_50sim_fixpoints_bliss_modelwise_synergies.tab")
topolink_ss_bliss_ew_150sim_file = paste0("results/topo-and-link/cascade_2.0_ss_150sim_fixpoints_bliss_ensemblewise_synergies.tab")
topolink_ss_bliss_mw_150sim_file = paste0("results/topo-and-link/cascade_2.0_ss_150sim_fixpoints_bliss_modelwise_synergies.tab")
topolink_ss_bliss_ew_synergies_50sim = emba::get_synergy_scores(topolink_ss_bliss_ew_50sim_file)
topolink_ss_bliss_mw_synergies_50sim = emba::get_synergy_scores(topolink_ss_bliss_mw_50sim_file, file_type = "modelwise")
topolink_ss_bliss_ew_synergies_150sim = emba::get_synergy_scores(topolink_ss_bliss_ew_150sim_file)
topolink_ss_bliss_mw_synergies_150sim = emba::get_synergy_scores(topolink_ss_bliss_mw_150sim_file, file_type = "modelwise")
## Bliss results rand
topolink_prolif_bliss_ew_50sim_file = paste0("results/topo-and-link/cascade_2.0_rand_50sim_fixpoints_bliss_ensemblewise_synergies.tab")
topolink_prolif_bliss_mw_50sim_file = paste0("results/topo-and-link/cascade_2.0_rand_50sim_fixpoints_bliss_modelwise_synergies.tab")
topolink_prolif_bliss_ew_150sim_file = paste0("results/topo-and-link/cascade_2.0_rand_150sim_fixpoints_bliss_ensemblewise_synergies.tab")
topolink_prolif_bliss_mw_150sim_file = paste0("results/topo-and-link/cascade_2.0_rand_150sim_fixpoints_bliss_modelwise_synergies.tab")
topolink_prolif_bliss_ew_synergies_50sim = emba::get_synergy_scores(topolink_prolif_bliss_ew_50sim_file)
topolink_prolif_bliss_mw_synergies_50sim = emba::get_synergy_scores(topolink_prolif_bliss_mw_50sim_file, file_type = "modelwise")
topolink_prolif_bliss_ew_synergies_150sim = emba::get_synergy_scores(topolink_prolif_bliss_ew_150sim_file)
topolink_prolif_bliss_mw_synergies_150sim = emba::get_synergy_scores(topolink_prolif_bliss_mw_150sim_file, file_type = "modelwise")
# calculate probability of synergy in the modelwise results
topolink_ss_hsa_mw_synergies_50sim = topolink_ss_hsa_mw_synergies_50sim %>%
mutate(synergy_prob_ss = synergies/(synergies + `non-synergies`))
topolink_ss_hsa_mw_synergies_150sim = topolink_ss_hsa_mw_synergies_150sim %>%
mutate(synergy_prob_ss = synergies/(synergies + `non-synergies`))
topolink_prolif_hsa_mw_synergies_50sim = topolink_prolif_hsa_mw_synergies_50sim %>%
mutate(synergy_prob_ss = synergies/(synergies + `non-synergies`))
topolink_prolif_hsa_mw_synergies_150sim = topolink_prolif_hsa_mw_synergies_150sim %>%
mutate(synergy_prob_ss = synergies/(synergies + `non-synergies`))
topolink_ss_bliss_mw_synergies_50sim = topolink_ss_bliss_mw_synergies_50sim %>%
mutate(synergy_prob_ss = synergies/(synergies + `non-synergies`))
topolink_ss_bliss_mw_synergies_150sim = topolink_ss_bliss_mw_synergies_150sim %>%
mutate(synergy_prob_ss = synergies/(synergies + `non-synergies`))
topolink_prolif_bliss_mw_synergies_50sim = topolink_prolif_bliss_mw_synergies_50sim %>%
mutate(synergy_prob_ss = synergies/(synergies + `non-synergies`))
topolink_prolif_bliss_mw_synergies_150sim = topolink_prolif_bliss_mw_synergies_150sim %>%
mutate(synergy_prob_ss = synergies/(synergies + `non-synergies`))
# Tidy the data
pred_topolink_ew_hsa = bind_cols(
topolink_ss_hsa_ew_synergies_50sim %>% rename(ss_score_50sim = score),
topolink_ss_hsa_ew_synergies_150sim %>% select(score) %>% rename(ss_score_150sim = score),
topolink_prolif_hsa_ew_synergies_50sim %>% select(score) %>% rename(prolif_score_50sim = score),
topolink_prolif_hsa_ew_synergies_150sim %>% select(score) %>% rename(prolif_score_150sim = score),
as_tibble_col(observed, column_name = "observed"))
pred_topolink_mw_hsa = bind_cols(
topolink_ss_hsa_mw_synergies_50sim %>% select(perturbation, synergy_prob_ss) %>% rename(synergy_prob_ss_50sim = synergy_prob_ss),
topolink_ss_hsa_mw_synergies_150sim %>% select(synergy_prob_ss) %>% rename(synergy_prob_ss_150sim = synergy_prob_ss),
topolink_prolif_hsa_mw_synergies_50sim %>% select(synergy_prob_ss) %>% rename(synergy_prob_prolif_50sim = synergy_prob_ss),
topolink_prolif_hsa_mw_synergies_150sim %>% select(synergy_prob_ss) %>% rename(synergy_prob_prolif_150sim = synergy_prob_ss),
as_tibble_col(observed, column_name = "observed"))
pred_topolink_ew_bliss = bind_cols(
topolink_ss_bliss_ew_synergies_50sim %>% rename(ss_score_50sim = score),
topolink_ss_bliss_ew_synergies_150sim %>% select(score) %>% rename(ss_score_150sim = score),
topolink_prolif_bliss_ew_synergies_50sim %>% select(score) %>% rename(prolif_score_50sim = score),
topolink_prolif_bliss_ew_synergies_150sim %>% select(score) %>% rename(prolif_score_150sim = score),
as_tibble_col(observed, column_name = "observed"))
pred_topolink_mw_bliss = bind_cols(
topolink_ss_bliss_mw_synergies_50sim %>% select(perturbation, synergy_prob_ss) %>% rename(synergy_prob_ss_50sim = synergy_prob_ss),
topolink_ss_bliss_mw_synergies_150sim %>% select(synergy_prob_ss) %>% rename(synergy_prob_ss_150sim = synergy_prob_ss),
topolink_prolif_bliss_mw_synergies_50sim %>% select(synergy_prob_ss) %>% rename(synergy_prob_prolif_50sim = synergy_prob_ss),
topolink_prolif_bliss_mw_synergies_150sim %>% select(synergy_prob_ss) %>% rename(synergy_prob_prolif_150sim = synergy_prob_ss),
as_tibble_col(observed, column_name = "observed"))HSA Results
- HSA refers to the synergy method used in
Drabmeto assess the synergies from thegitsbemodels - We test performance using ROC and PR AUC for both the ensemble-wise and model-wise synergies from
Drabme - Calibrated models: fitted to steady state (\(50,150\) simulations)
- Random models: fitted to proliferation profile (\(50,150\) simulations)
Gitsbemodels have both balance and topology mutations (\(3000,50\) mutations as a bootstrap value, \(3\) and \(10\) respectively after models with stable states are found)
ROC curves
topolink_res_ss_ew_50sim = get_roc_stats(df = pred_topolink_ew_hsa, pred_col = "ss_score_50sim", label_col = "observed")
topolink_res_ss_ew_150sim = get_roc_stats(df = pred_topolink_ew_hsa, pred_col = "ss_score_150sim", label_col = "observed")
topolink_res_prolif_ew_50sim = get_roc_stats(df = pred_topolink_ew_hsa, pred_col = "prolif_score_50sim", label_col = "observed")
topolink_res_prolif_ew_150sim = get_roc_stats(df = pred_topolink_ew_hsa, pred_col = "prolif_score_150sim", label_col = "observed")
topolink_res_ss_mw_50sim = get_roc_stats(df = pred_topolink_mw_hsa, pred_col = "synergy_prob_ss_50sim", label_col = "observed", direction = ">")
topolink_res_ss_mw_150sim = get_roc_stats(df = pred_topolink_mw_hsa, pred_col = "synergy_prob_ss_150sim", label_col = "observed", direction = ">")
topolink_res_prolif_mw_50sim = get_roc_stats(df = pred_topolink_mw_hsa, pred_col = "synergy_prob_prolif_50sim", label_col = "observed", direction = ">")
topolink_res_prolif_mw_150sim = get_roc_stats(df = pred_topolink_mw_hsa, pred_col = "synergy_prob_prolif_150sim", label_col = "observed", direction = ">")
# Plot ROCs
plot(x = topolink_res_ss_ew_50sim$roc_stats$FPR, y = topolink_res_ss_ew_50sim$roc_stats$TPR,
type = 'l', lwd = 3, col = my_palette[1], main = 'ROC curve, Ensemble-wise synergies (HSA)',
xlab = 'False Positive Rate (FPR)', ylab = 'True Positive Rate (TPR)')
lines(x = topolink_res_ss_ew_150sim$roc_stats$FPR, y = topolink_res_ss_ew_150sim$roc_stats$TPR,
lwd = 3, col = my_palette[2])
lines(x = topolink_res_prolif_ew_50sim$roc_stats$FPR, y = topolink_res_prolif_ew_50sim$roc_stats$TPR,
lwd = 3, col = my_palette[3])
lines(x = topolink_res_prolif_ew_150sim$roc_stats$FPR, y = topolink_res_prolif_ew_150sim$roc_stats$TPR,
lwd = 3, col = my_palette[4])
legend('bottomright', title = 'AUC', col = my_palette[1:4], pch = 19,
legend = c(paste(round(topolink_res_ss_ew_50sim$AUC, digits = 2), "Calibrated (50 sim)"),
paste(round(topolink_res_ss_ew_150sim$AUC, digits = 2), "Calibrated (150 sim)"),
paste(round(topolink_res_prolif_ew_50sim$AUC, digits = 2), "Random (50 sim)"),
paste(round(topolink_res_prolif_ew_150sim$AUC, digits = 2), "Random (150 sim)")))
grid(lwd = 0.5)
abline(a = 0, b = 1, col = 'lightgrey', lty = 'dotdash', lwd = 1.2)
plot(x = topolink_res_ss_mw_50sim$roc_stats$FPR, y = topolink_res_ss_mw_50sim$roc_stats$TPR,
type = 'l', lwd = 3, col = my_palette[1], main = 'ROC curve, Model-wise synergies (HSA)',
xlab = 'False Positive Rate (FPR)', ylab = 'True Positive Rate (TPR)')
lines(x = topolink_res_ss_mw_150sim$roc_stats$FPR, y = topolink_res_ss_mw_150sim$roc_stats$TPR,
lwd = 3, col = my_palette[2])
lines(x = topolink_res_prolif_mw_50sim$roc_stats$FPR, y = topolink_res_prolif_mw_50sim$roc_stats$TPR,
lwd = 3, col = my_palette[3])
lines(x = topolink_res_prolif_mw_150sim$roc_stats$FPR, y = topolink_res_prolif_mw_150sim$roc_stats$TPR,
lwd = 3, col = my_palette[4])
legend('bottomright', title = 'AUC', col = my_palette[1:4], pch = 19,
legend = c(paste(round(topolink_res_ss_mw_50sim$AUC, digits = 2), "Calibrated (50 sim)"),
paste(round(topolink_res_ss_mw_150sim$AUC, digits = 2), "Calibrated (150 sim)"),
paste(round(topolink_res_prolif_mw_50sim$AUC, digits = 2), "Random (50 sim)"),
paste(round(topolink_res_prolif_mw_150sim$AUC, digits = 2), "Random (150 sim)")))
grid(lwd = 0.5)
abline(a = 0, b = 1, col = 'lightgrey', lty = 'dotdash', lwd = 1.2)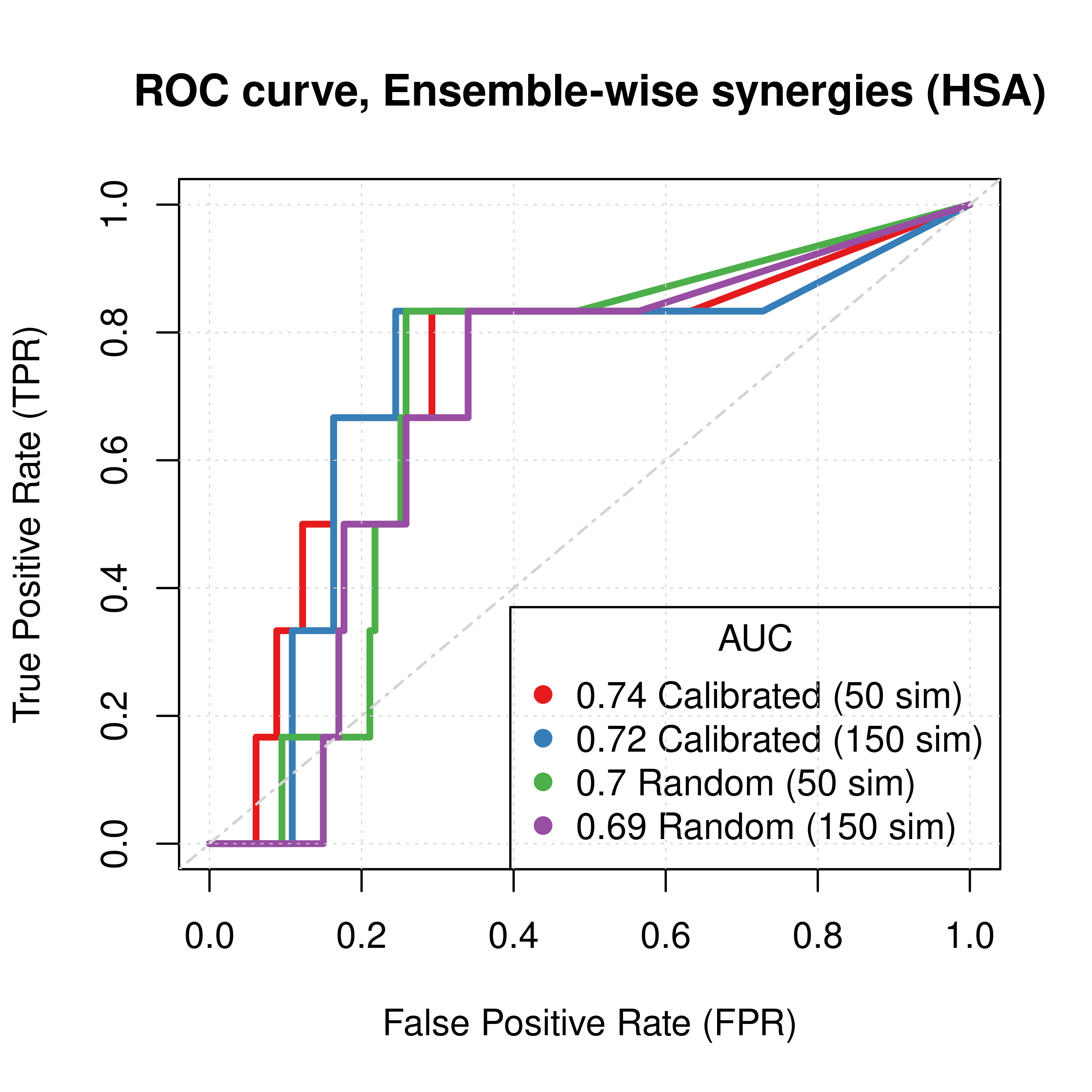
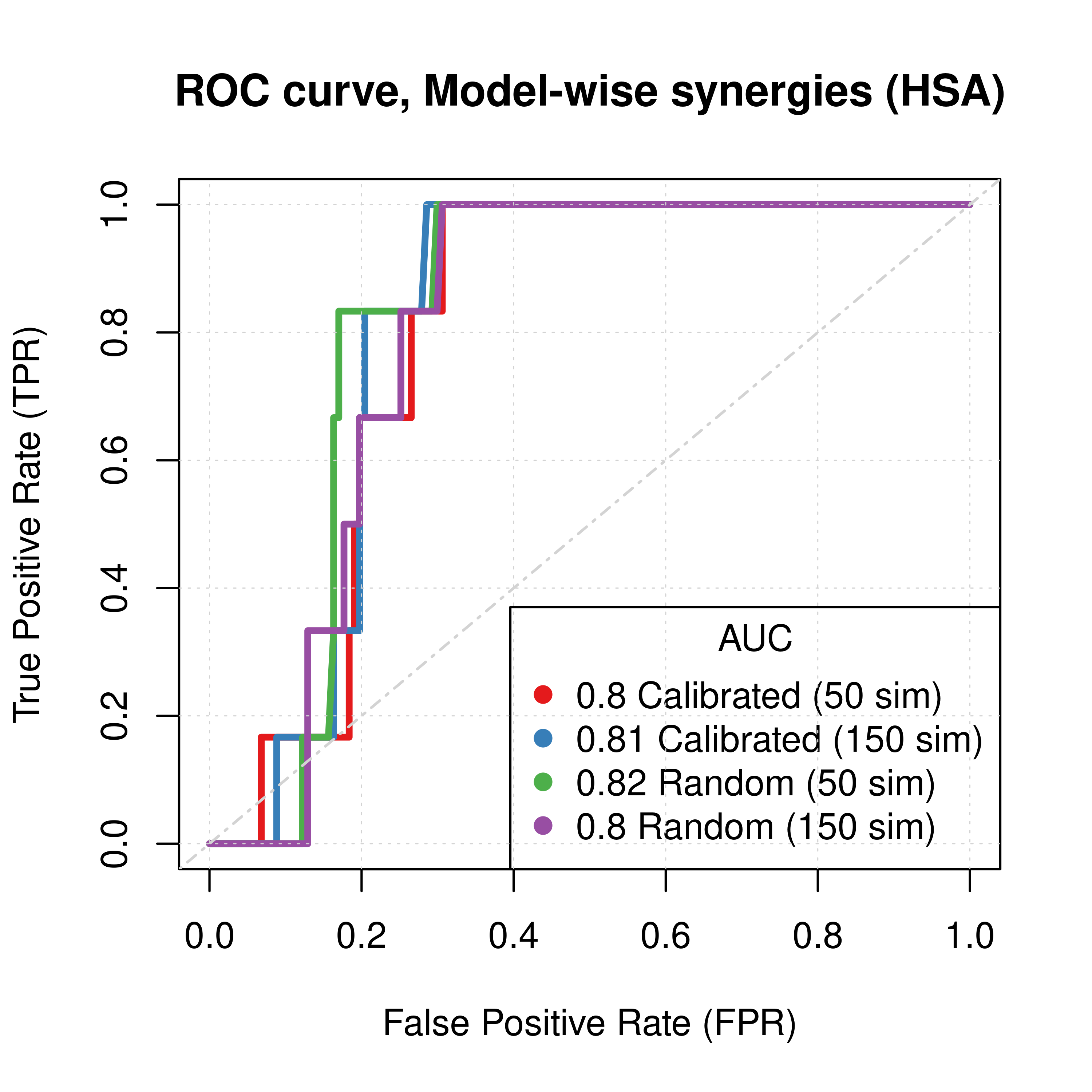
Figure 58: ROC curves (CASCADE 2.0, Link Operator and Topology Mutations, HSA synergy method)
PR curves
pr_topolink_res_ss_ew_50sim = pr.curve(scores.class0 = pred_topolink_ew_hsa %>% pull(ss_score_50sim) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_hsa %>% pull(observed), curve = TRUE, rand.compute = TRUE)
pr_topolink_res_ss_ew_150sim = pr.curve(scores.class0 = pred_topolink_ew_hsa %>% pull(ss_score_150sim) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_hsa %>% pull(observed), curve = TRUE)
pr_topolink_res_prolif_ew_50sim = pr.curve(scores.class0 = pred_topolink_ew_hsa %>% pull(prolif_score_50sim) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_hsa %>% pull(observed), curve = TRUE)
pr_topolink_res_prolif_ew_150sim = pr.curve(scores.class0 = pred_topolink_ew_hsa %>% pull(prolif_score_150sim) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_hsa %>% pull(observed), curve = TRUE)
pr_topolink_res_ss_mw_50sim = pr.curve(scores.class0 = pred_topolink_mw_hsa %>% pull(synergy_prob_ss_50sim),
weights.class0 = pred_topolink_mw_hsa %>% pull(observed), curve = TRUE, rand.compute = TRUE)
pr_topolink_res_ss_mw_150sim = pr.curve(scores.class0 = pred_topolink_mw_hsa %>% pull(synergy_prob_ss_150sim),
weights.class0 = pred_topolink_mw_hsa %>% pull(observed), curve = TRUE)
pr_topolink_res_prolif_mw_50sim = pr.curve(scores.class0 = pred_topolink_mw_hsa %>% pull(synergy_prob_prolif_50sim),
weights.class0 = pred_topolink_mw_hsa %>% pull(observed), curve = TRUE)
pr_topolink_res_prolif_mw_150sim = pr.curve(scores.class0 = pred_topolink_mw_hsa %>% pull(synergy_prob_prolif_150sim),
weights.class0 = pred_topolink_mw_hsa %>% pull(observed), curve = TRUE)
plot(pr_topolink_res_ss_ew_50sim, main = 'PR curve, Ensemble-wise synergies (HSA)',
auc.main = FALSE, color = my_palette[1], rand.plot = TRUE)
plot(pr_topolink_res_ss_ew_150sim, add = TRUE, color = my_palette[2])
plot(pr_topolink_res_prolif_ew_50sim, add = TRUE, color = my_palette[3])
plot(pr_topolink_res_prolif_ew_150sim, add = TRUE, color = my_palette[4])
legend('topright', title = 'AUC', col = my_palette[1:4], pch = 19,
legend = c(paste(round(pr_topolink_res_ss_ew_50sim$auc.davis.goadrich, digits = 3), "Calibrated (50 sim)"),
paste(round(pr_topolink_res_ss_ew_150sim$auc.davis.goadrich, digits = 3), "Calibrated (150 sim)"),
paste(round(pr_topolink_res_prolif_ew_50sim$auc.davis.goadrich, digits = 3), "Random (50 sim)"),
paste(round(pr_topolink_res_prolif_ew_150sim$auc.davis.goadrich, digits = 3), "Random (150 sim)")))
grid(lwd = 0.5)
plot(pr_topolink_res_ss_mw_50sim, main = 'PR curve, Model-wise synergies (HSA)',
auc.main = FALSE, color = my_palette[1], rand.plot = TRUE)
plot(pr_topolink_res_ss_mw_150sim, add = TRUE, color = my_palette[2])
plot(pr_topolink_res_prolif_mw_50sim, add = TRUE, color = my_palette[3])
plot(pr_topolink_res_prolif_mw_150sim, add = TRUE, color = my_palette[4])
legend('topright', title = 'AUC', col = my_palette[1:4], pch = 19,
legend = c(paste(round(pr_topolink_res_ss_mw_50sim$auc.davis.goadrich, digits = 3), "Calibrated (50 sim)"),
paste(round(pr_topolink_res_ss_mw_150sim$auc.davis.goadrich, digits = 3), "Calibrated (150 sim)"),
paste(round(pr_topolink_res_prolif_mw_50sim$auc.davis.goadrich, digits = 3), "Random (50 sim)"),
paste(round(pr_topolink_res_prolif_mw_150sim$auc.davis.goadrich, digits = 3), "Random (150 sim)")))
grid(lwd = 0.5)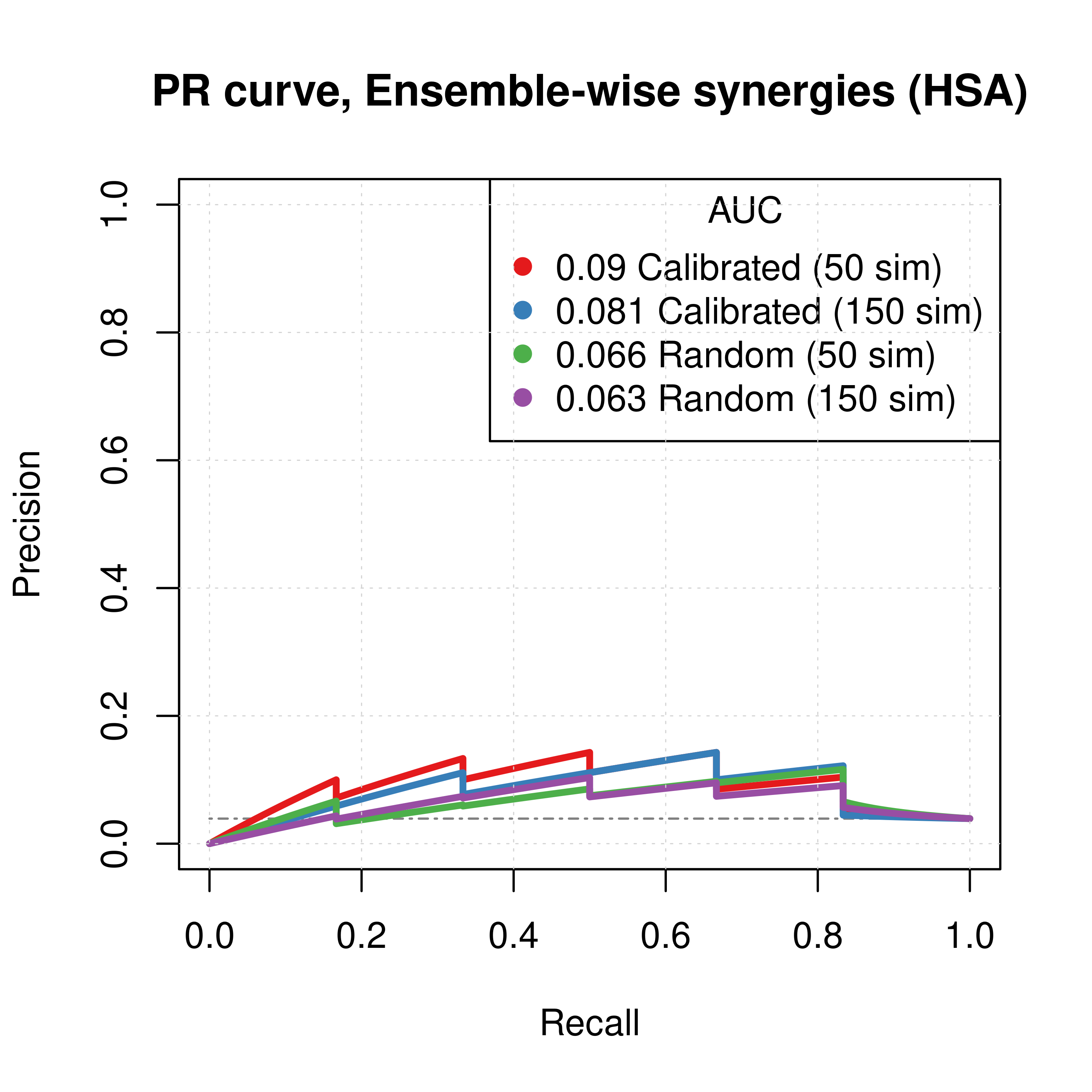
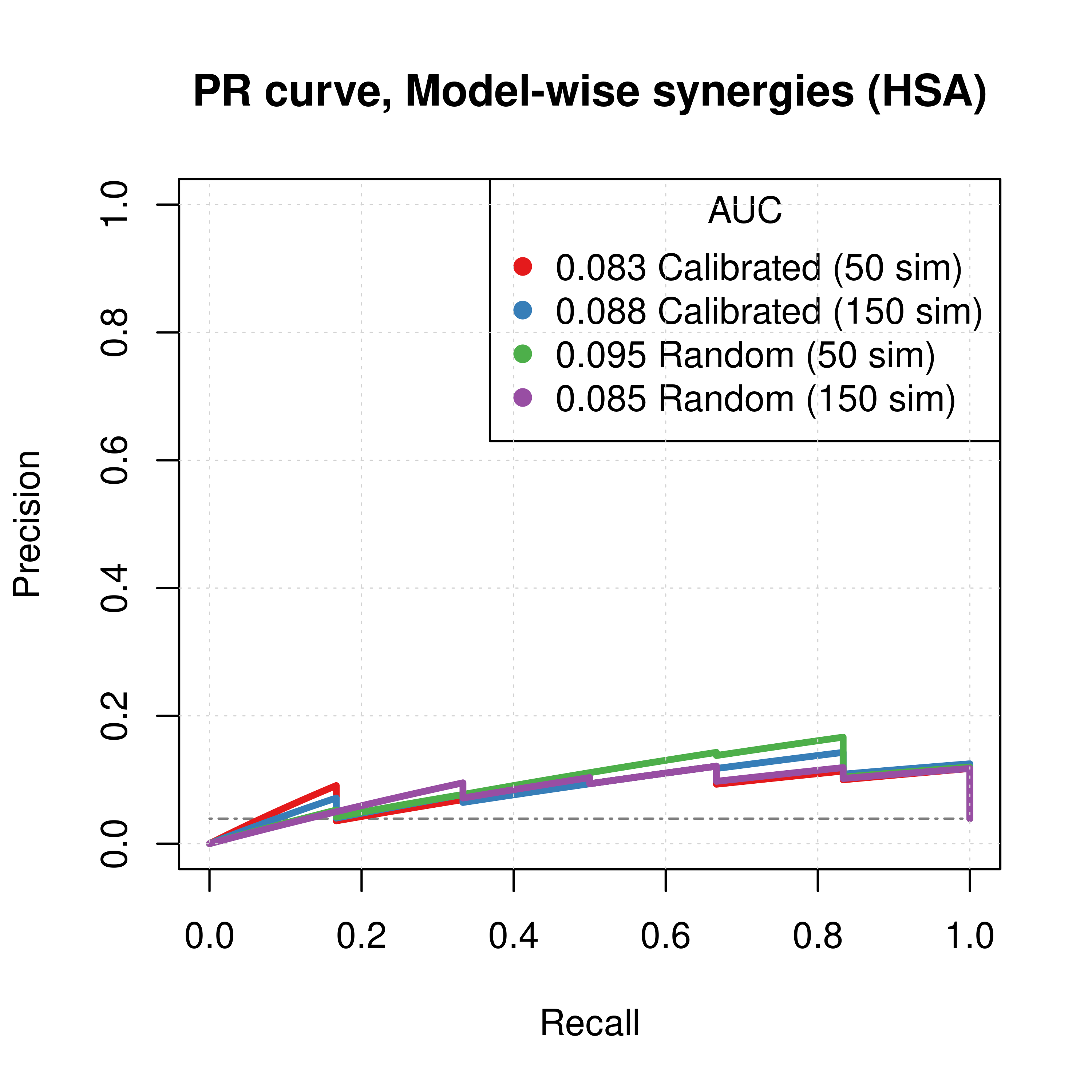
Figure 59: PR curves (CASCADE 2.0, Link Operator and Topology Mutations, HSA synergy method)
- The PR curves show that the performance of each individual predictor is poor compared to the baseline. Someone looking at the ROC curves only might reach a different conclusion.
- The model-wise approach produces slightly better ROC results than the ensemble-wise approach.
AUC sensitivity
Investigate same thing as described in here. This is very crucial since the PR performance is poor for the individual predictors, but a combined predictor might be able to counter this. We will combine the synergy scores from the random proliferative simulations with the results from the calibrated Gitsbe simulations (number of simulations: \(150\)).
# Ensemble-wise
betas = seq(from = -5, to = 5, by = 0.1)
prolif_roc_topo = sapply(betas, function(beta) {
pred_topolink_ew_hsa = pred_topolink_ew_hsa %>% mutate(combined_score = ss_score_150sim + beta * prolif_score_150sim)
res = roc.curve(scores.class0 = pred_topolink_ew_hsa %>% pull(combined_score) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_hsa %>% pull(observed))
auc_value = res$auc
})
prolif_pr_topo = sapply(betas, function(beta) {
pred_topolink_ew_hsa = pred_topolink_ew_hsa %>% mutate(combined_score = ss_score_150sim + beta * prolif_score_150sim)
res = pr.curve(scores.class0 = pred_topolink_ew_hsa %>% pull(combined_score) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_hsa %>% pull(observed))
auc_value = res$auc.davis.goadrich
})
df_ew = as_tibble(cbind(betas, prolif_roc_topo, prolif_pr_topo))
df_ew = df_ew %>% tidyr::pivot_longer(-betas, names_to = "type", values_to = "AUC")
ggline(data = df_ew, x = "betas", y = "AUC", numeric.x.axis = TRUE, color = "type",
plot_type = "l", xlab = TeX("$\\beta$"), ylab = "AUC (Area Under Curve)",
legend = "none", facet.by = "type", palette = my_palette, ylim = c(0,0.85),
panel.labs = list(type = c("Precision-Recall", "ROC")),
title = TeX("AUC sensitivity to $\\beta$ parameter")) +
theme(plot.title = element_text(hjust = 0.5)) +
geom_vline(xintercept = 0) +
geom_vline(xintercept = -1, color = "black", size = 0.3, linetype = "dashed") +
geom_text(aes(x=-1.6, label="β = -1", y=0.33), colour="black", angle=90) +
grids()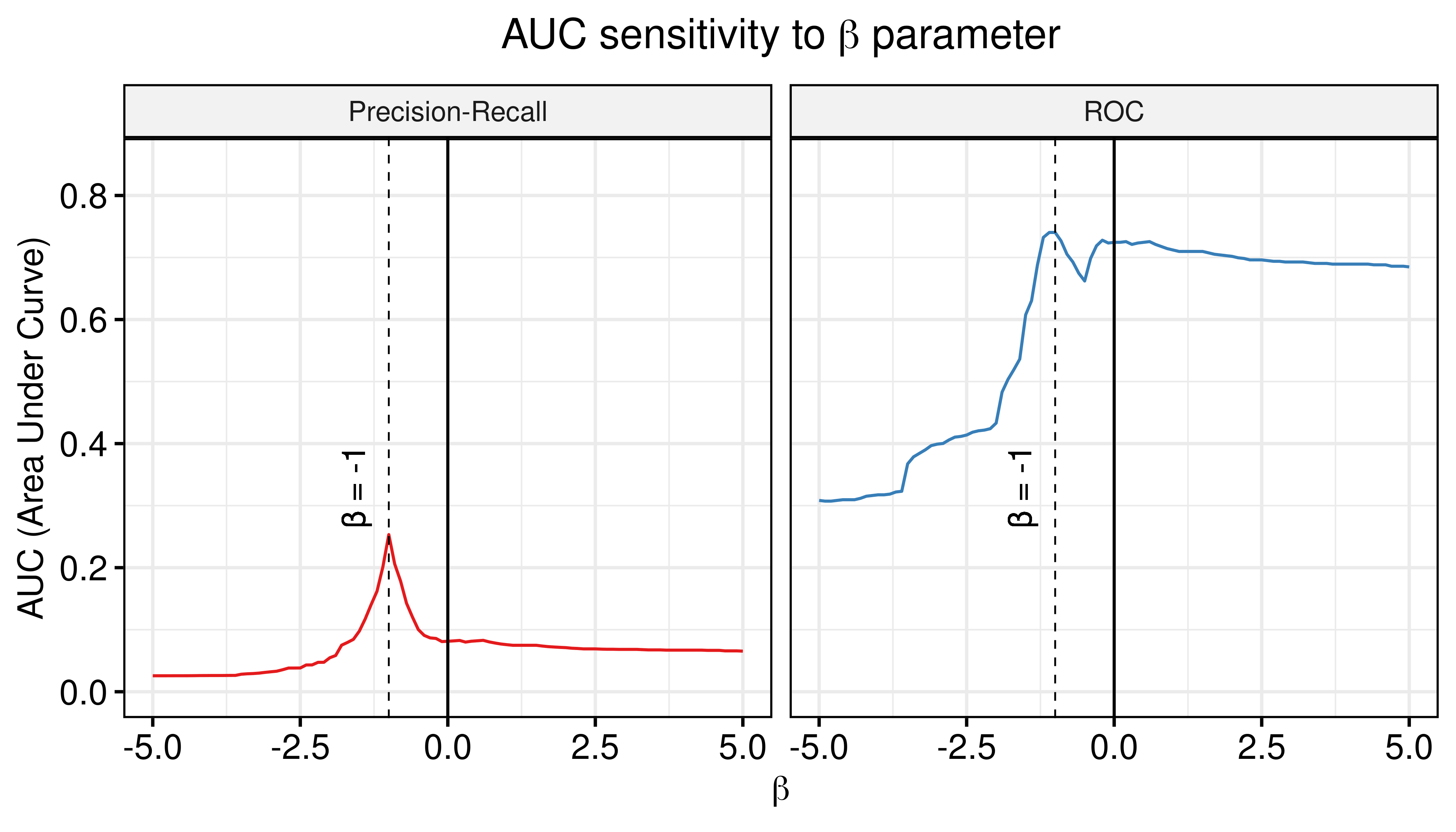
Figure 60: AUC sensitivity (CASCADE 2.0, Link Operator and Topology Mutations, HSA synergy method, Ensemble-wise results)
- The random proliferative models can be used to normalize against the predictions of the calibrated models and thus bring significant contribution to the calibrated models performance (PR-AUC shows much more sensitivity in that regard - it increases substantially more than the ROC-AUC).
- The \(\beta_{best}\) value of the combined calibrated and random proliferative model predictor that maximizes both the ROC-AUC and PR-AUC is \(\beta_{best}=-1\).
Bliss Results
- Bliss refers to the synergy method used in
Drabmeto assess the synergies from thegitsbemodels - We test performance using ROC and PR AUC for both the ensemble-wise and model-wise synergies from
Drabme - Calibrated models: fitted to steady state (\(50,150\) simulations)
- Random models: fitted to proliferation profile (\(50,150\) simulations)
Gitsbemodels have both balance and topology mutations (\(3000,50\) mutations as a bootstrap value, \(3\) and \(10\) respectively after models with stable states are found)
ROC curves
topolink_res_ss_ew_50sim = get_roc_stats(df = pred_topolink_ew_bliss, pred_col = "ss_score_50sim", label_col = "observed")
topolink_res_ss_ew_150sim = get_roc_stats(df = pred_topolink_ew_bliss, pred_col = "ss_score_150sim", label_col = "observed")
topolink_res_prolif_ew_50sim = get_roc_stats(df = pred_topolink_ew_bliss, pred_col = "prolif_score_50sim", label_col = "observed")
topolink_res_prolif_ew_150sim = get_roc_stats(df = pred_topolink_ew_bliss, pred_col = "prolif_score_150sim", label_col = "observed")
topolink_res_ss_mw_50sim = get_roc_stats(df = pred_topolink_mw_bliss, pred_col = "synergy_prob_ss_50sim", label_col = "observed", direction = ">")
topolink_res_ss_mw_150sim = get_roc_stats(df = pred_topolink_mw_bliss, pred_col = "synergy_prob_ss_150sim", label_col = "observed", direction = ">")
topolink_res_prolif_mw_50sim = get_roc_stats(df = pred_topolink_mw_bliss, pred_col = "synergy_prob_prolif_50sim", label_col = "observed", direction = ">")
topolink_res_prolif_mw_150sim = get_roc_stats(df = pred_topolink_mw_bliss, pred_col = "synergy_prob_prolif_150sim", label_col = "observed", direction = ">")
# Plot ROCs
plot(x = topolink_res_ss_ew_50sim$roc_stats$FPR, y = topolink_res_ss_ew_50sim$roc_stats$TPR,
type = 'l', lwd = 3, col = my_palette[1], main = 'ROC curve, Ensemble-wise synergies (Bliss)',
xlab = 'False Positive Rate (FPR)', ylab = 'True Positive Rate (TPR)')
lines(x = topolink_res_ss_ew_150sim$roc_stats$FPR, y = topolink_res_ss_ew_150sim$roc_stats$TPR,
lwd = 3, col = my_palette[2])
lines(x = topolink_res_prolif_ew_50sim$roc_stats$FPR, y = topolink_res_prolif_ew_50sim$roc_stats$TPR,
lwd = 3, col = my_palette[3])
lines(x = topolink_res_prolif_ew_150sim$roc_stats$FPR, y = topolink_res_prolif_ew_150sim$roc_stats$TPR,
lwd = 3, col = my_palette[4])
legend('bottomright', title = 'AUC', col = my_palette[1:4], pch = 19,
legend = c(paste(round(topolink_res_ss_ew_50sim$AUC, digits = 2), "Calibrated (50 sim)"),
paste(round(topolink_res_ss_ew_150sim$AUC, digits = 2), "Calibrated (150 sim)"),
paste(round(topolink_res_prolif_ew_50sim$AUC, digits = 2), "Random (50 sim)"),
paste(round(topolink_res_prolif_ew_150sim$AUC, digits = 2), "Random (150 sim)")))
grid(lwd = 0.5)
abline(a = 0, b = 1, col = 'lightgrey', lty = 'dotdash', lwd = 1.2)
plot(x = topolink_res_ss_mw_50sim$roc_stats$FPR, y = topolink_res_ss_mw_50sim$roc_stats$TPR,
type = 'l', lwd = 3, col = my_palette[1], main = 'ROC curve, Model-wise synergies (Bliss)',
xlab = 'False Positive Rate (FPR)', ylab = 'True Positive Rate (TPR)')
lines(x = topolink_res_ss_mw_150sim$roc_stats$FPR, y = topolink_res_ss_mw_150sim$roc_stats$TPR,
lwd = 3, col = my_palette[2])
lines(x = topolink_res_prolif_mw_50sim$roc_stats$FPR, y = topolink_res_prolif_mw_50sim$roc_stats$TPR,
lwd = 3, col = my_palette[3])
lines(x = topolink_res_prolif_mw_150sim$roc_stats$FPR, y = topolink_res_prolif_mw_150sim$roc_stats$TPR,
lwd = 3, col = my_palette[4])
legend('bottomright', title = 'AUC', col = my_palette[1:4], pch = 19,
legend = c(paste(round(topolink_res_ss_mw_50sim$AUC, digits = 2), "Calibrated (50 sim)"),
paste(round(topolink_res_ss_mw_150sim$AUC, digits = 2), "Calibrated (150 sim)"),
paste(round(topolink_res_prolif_mw_50sim$AUC, digits = 2), "Random (50 sim)"),
paste(round(topolink_res_prolif_mw_150sim$AUC, digits = 2), "Random (150 sim)")))
grid(lwd = 0.5)
abline(a = 0, b = 1, col = 'lightgrey', lty = 'dotdash', lwd = 1.2)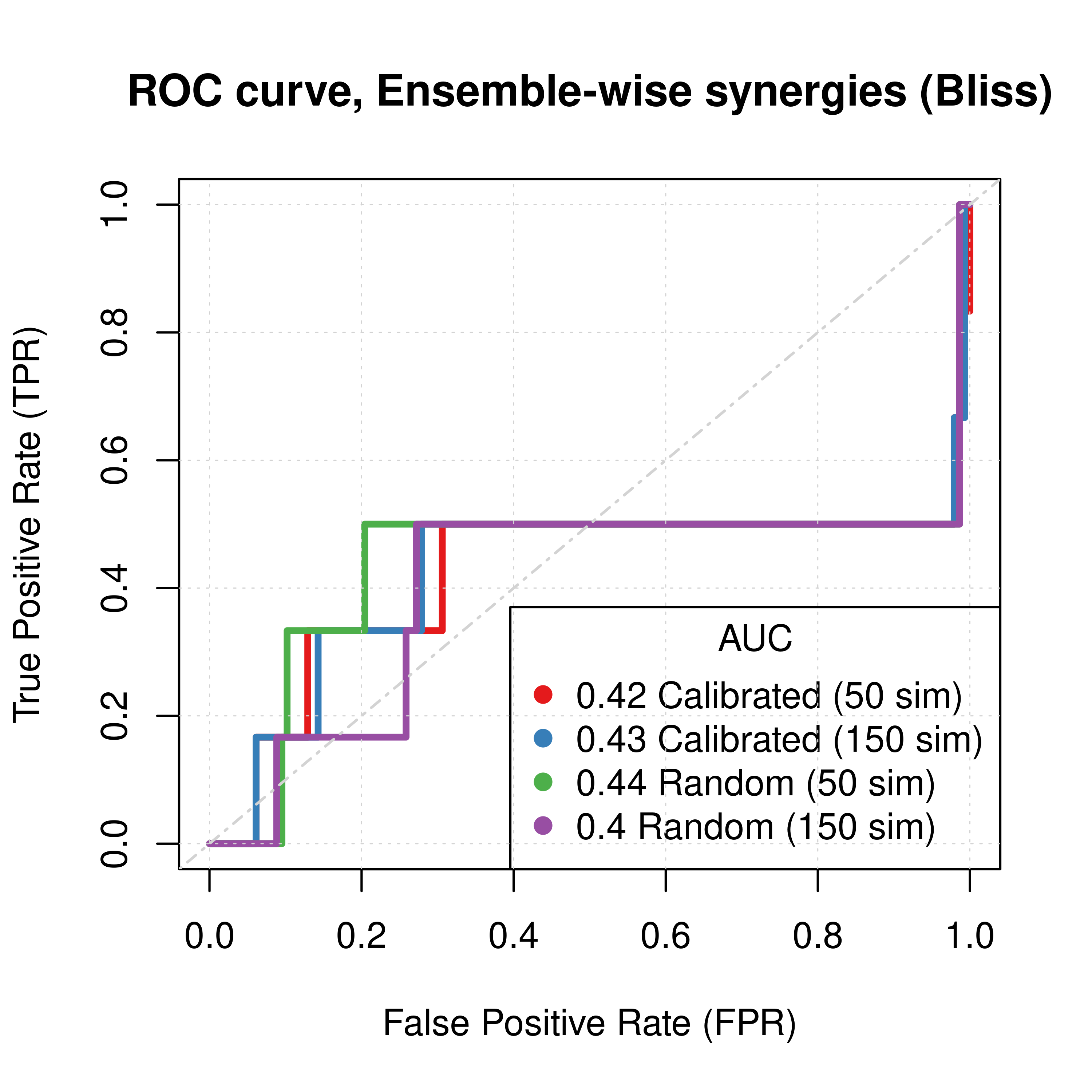
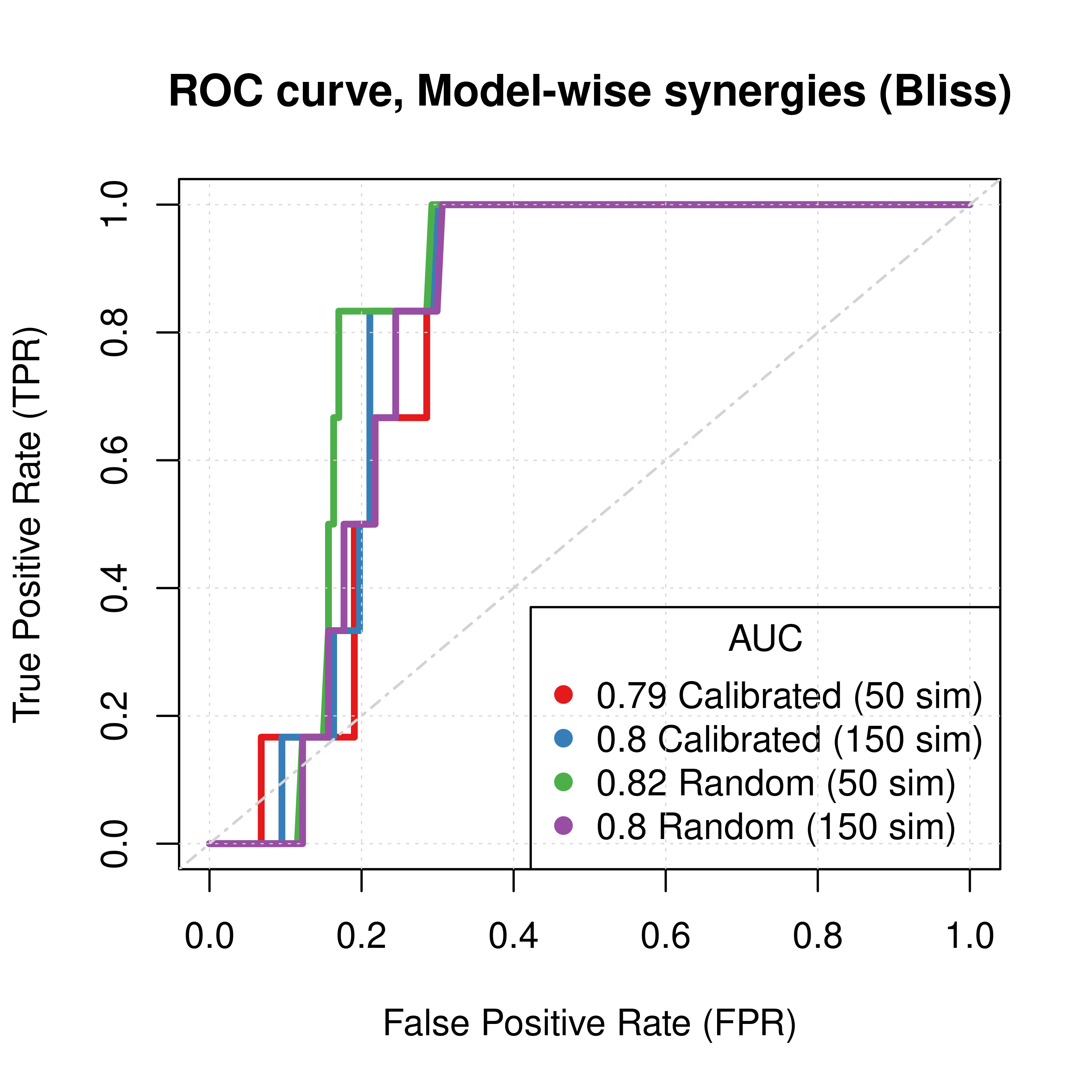
Figure 61: ROC curves (CASCADE 2.0, Link Operator and Topology Mutations, Bliss synergy method)
PR curves
pr_topolink_res_ss_ew_50sim = pr.curve(scores.class0 = pred_topolink_ew_bliss %>% pull(ss_score_50sim) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_bliss %>% pull(observed), curve = TRUE, rand.compute = TRUE)
pr_topolink_res_ss_ew_150sim = pr.curve(scores.class0 = pred_topolink_ew_bliss %>% pull(ss_score_150sim) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_bliss %>% pull(observed), curve = TRUE)
pr_topolink_res_prolif_ew_50sim = pr.curve(scores.class0 = pred_topolink_ew_bliss %>% pull(prolif_score_50sim) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_bliss %>% pull(observed), curve = TRUE)
pr_topolink_res_prolif_ew_150sim = pr.curve(scores.class0 = pred_topolink_ew_bliss %>% pull(prolif_score_150sim) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_bliss %>% pull(observed), curve = TRUE)
pr_topolink_res_ss_mw_50sim = pr.curve(scores.class0 = pred_topolink_mw_bliss %>% pull(synergy_prob_ss_50sim),
weights.class0 = pred_topolink_mw_bliss %>% pull(observed), curve = TRUE, rand.compute = TRUE)
pr_topolink_res_ss_mw_150sim = pr.curve(scores.class0 = pred_topolink_mw_bliss %>% pull(synergy_prob_ss_150sim),
weights.class0 = pred_topolink_mw_bliss %>% pull(observed), curve = TRUE)
pr_topolink_res_prolif_mw_50sim = pr.curve(scores.class0 = pred_topolink_mw_bliss %>% pull(synergy_prob_prolif_50sim),
weights.class0 = pred_topolink_mw_bliss %>% pull(observed), curve = TRUE)
pr_topolink_res_prolif_mw_150sim = pr.curve(scores.class0 = pred_topolink_mw_bliss %>% pull(synergy_prob_prolif_150sim),
weights.class0 = pred_topolink_mw_bliss %>% pull(observed), curve = TRUE)
plot(pr_topolink_res_ss_ew_50sim, main = 'PR curve, Ensemble-wise synergies (Bliss)',
auc.main = FALSE, color = my_palette[1], rand.plot = TRUE)
plot(pr_topolink_res_ss_ew_150sim, add = TRUE, color = my_palette[2])
plot(pr_topolink_res_prolif_ew_50sim, add = TRUE, color = my_palette[3])
plot(pr_topolink_res_prolif_ew_150sim, add = TRUE, color = my_palette[4])
legend('topright', title = 'AUC', col = my_palette[1:4], pch = 19,
legend = c(paste(round(pr_topolink_res_ss_ew_50sim$auc.davis.goadrich, digits = 3), "Calibrated (50 sim)"),
paste(round(pr_topolink_res_ss_ew_150sim$auc.davis.goadrich, digits = 3), "Calibrated (150 sim)"),
paste(round(pr_topolink_res_prolif_ew_50sim$auc.davis.goadrich, digits = 3), "Random (50 sim)"),
paste(round(pr_topolink_res_prolif_ew_150sim$auc.davis.goadrich, digits = 3), "Random (150 sim)")))
grid(lwd = 0.5)
plot(pr_topolink_res_ss_mw_50sim, main = 'PR curve, Model-wise synergies (Bliss)',
auc.main = FALSE, color = my_palette[1], rand.plot = TRUE)
plot(pr_topolink_res_ss_mw_150sim, add = TRUE, color = my_palette[2])
plot(pr_topolink_res_prolif_mw_50sim, add = TRUE, color = my_palette[3])
plot(pr_topolink_res_prolif_mw_150sim, add = TRUE, color = my_palette[4])
legend('topright', title = 'AUC', col = my_palette[1:4], pch = 19,
legend = c(paste(round(pr_topolink_res_ss_mw_50sim$auc.davis.goadrich, digits = 3), "Calibrated (50 sim)"),
paste(round(pr_topolink_res_ss_mw_150sim$auc.davis.goadrich, digits = 3), "Calibrated (150 sim)"),
paste(round(pr_topolink_res_prolif_mw_50sim$auc.davis.goadrich, digits = 3), "Random (50 sim)"),
paste(round(pr_topolink_res_prolif_mw_150sim$auc.davis.goadrich, digits = 3), "Random (150 sim)")))
grid(lwd = 0.5)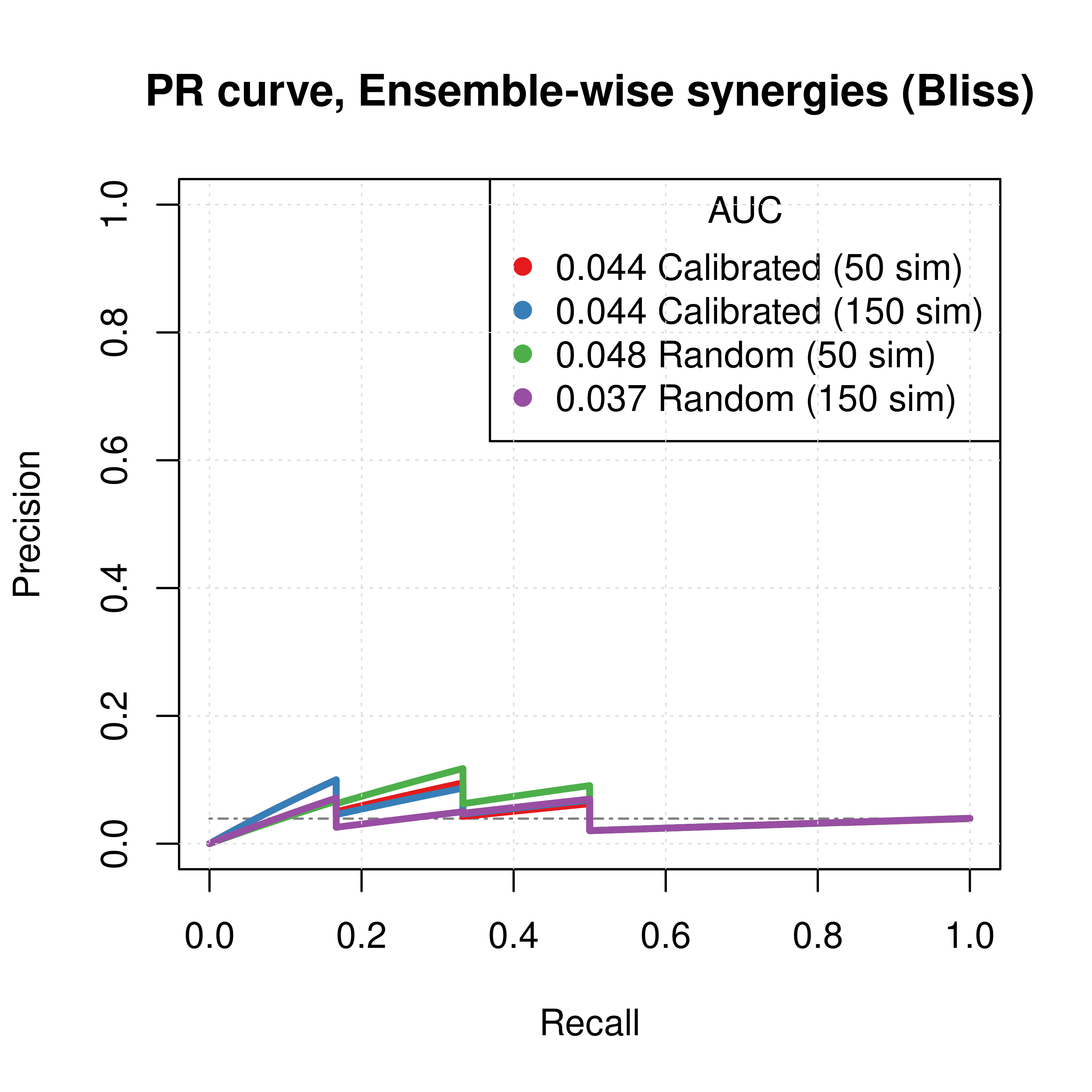
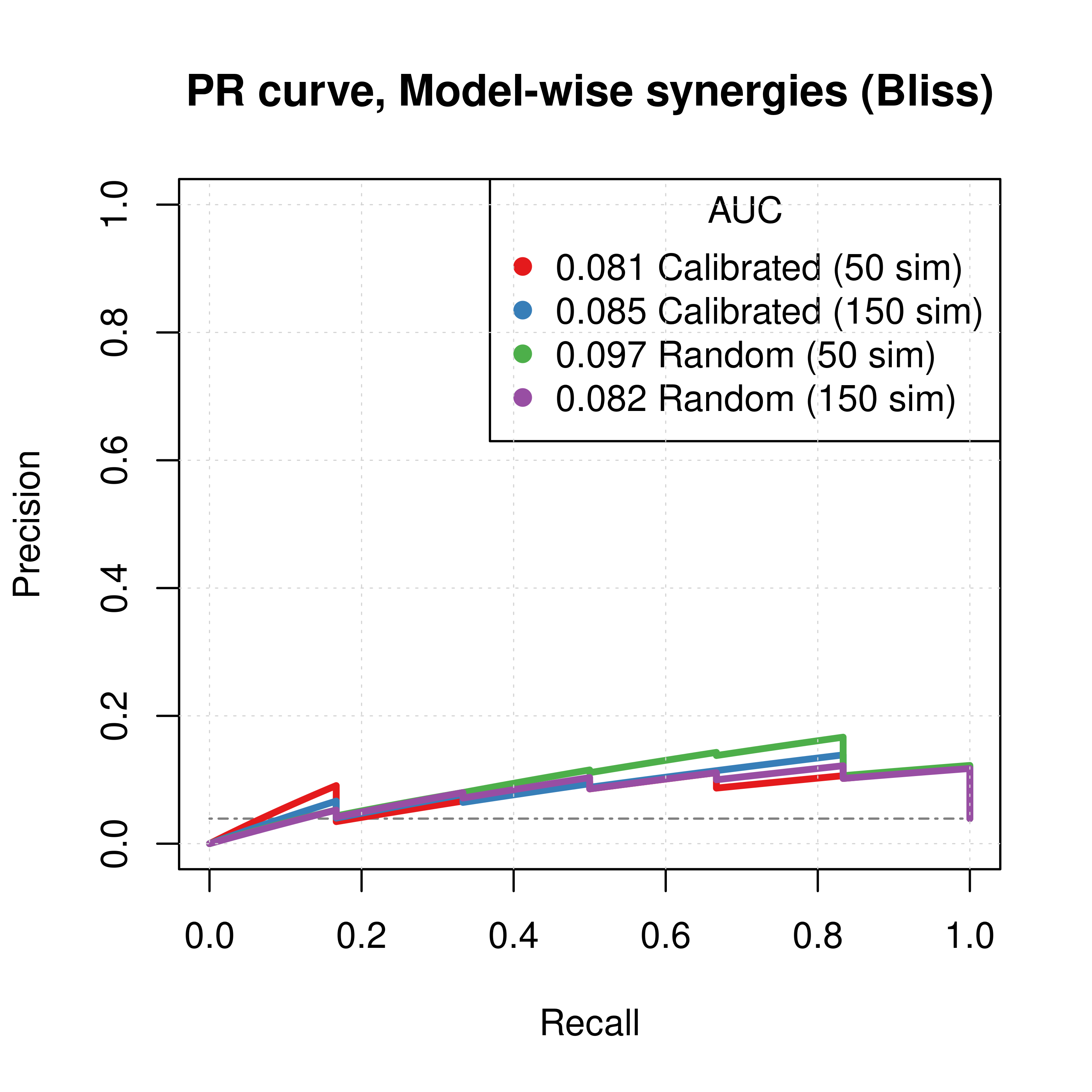
Figure 62: PR curves (CASCADE 2.0, Link Operator and Topology Mutations, Bliss synergy method)
- The PR curves show that the performance of each individual predictor is poor compared to the baseline.
- The model-wise approach produces better ROC and PR results than the ensemble-wise approach (performance in terms of AUC value is almost doubled)
AUC sensitivity
Investigate same thing as described in here. This is very crucial since the PR performance is poor for the individual predictors, but a combined predictor might be able to counter this. We will combine the synergy scores from the random proliferative simulations with the results from the calibrated Gitsbe simulations (number of simulations: \(150\)).
# Ensemble-wise
betas = seq(from = -5, to = 5, by = 0.1)
prolif_roc = sapply(betas, function(beta) {
pred_topolink_ew_bliss = pred_topolink_ew_bliss %>% mutate(combined_score = ss_score_150sim + beta * prolif_score_150sim)
res = roc.curve(scores.class0 = pred_topolink_ew_bliss %>% pull(combined_score) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_bliss %>% pull(observed))
auc_value = res$auc
})
prolif_pr = sapply(betas, function(beta) {
pred_topolink_ew_bliss = pred_topolink_ew_bliss %>% mutate(combined_score = ss_score_150sim + beta * prolif_score_150sim)
res = pr.curve(scores.class0 = pred_topolink_ew_bliss %>% pull(combined_score) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_bliss %>% pull(observed))
auc_value = res$auc.davis.goadrich
})
df_ew = as_tibble(cbind(betas, prolif_roc, prolif_pr))
df_ew = df_ew %>% tidyr::pivot_longer(-betas, names_to = "type", values_to = "AUC")
ggline(data = df_ew, x = "betas", y = "AUC", numeric.x.axis = TRUE, color = "type",
plot_type = "l", xlab = TeX("$\\beta$"), ylab = "AUC (Area Under Curve)",
legend = "none", facet.by = "type", palette = my_palette, ylim = c(0,0.85),
panel.labs = list(type = c("Precision-Recall", "ROC")),
title = TeX("AUC sensitivity to $\\beta$ parameter")) +
theme(plot.title = element_text(hjust = 0.5)) +
geom_vline(xintercept = 0) +
geom_vline(xintercept = -1, color = "black", size = 0.3, linetype = "dashed") +
geom_text(aes(x=-1.5, label="β = -1", y=0.35), colour="black", angle = 90) +
grids()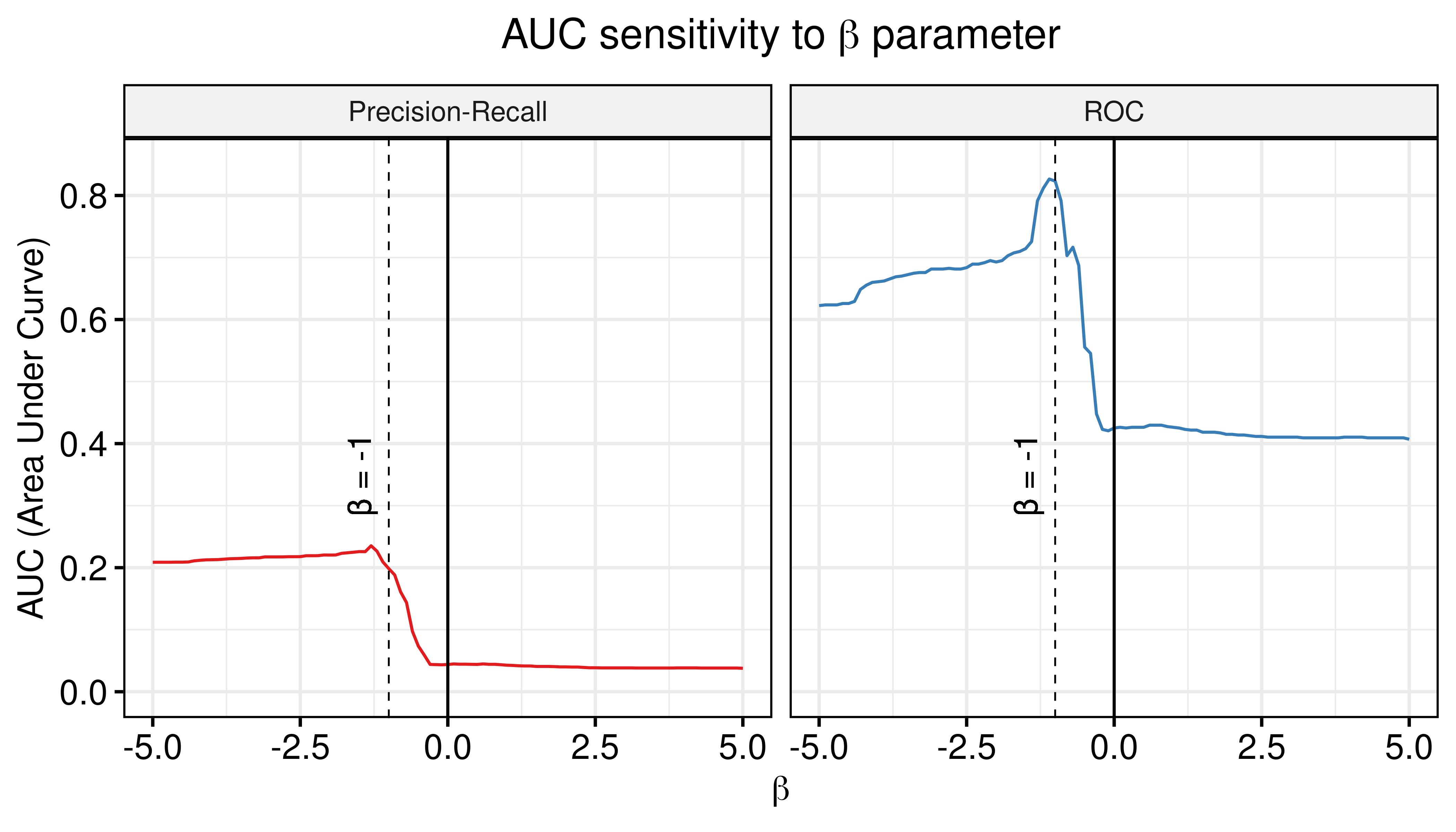
Figure 63: AUC sensitivity (CASCADE 2.0, Link Operator and Topology Mutations, Bliss synergy method, Ensemble-wise results)
- The random proliferative models can be used to normalize against the predictions of the calibrated models and thus bring significant contribution to the calibrated models performance (both ROC-AUC and PR-AUC are increased).
- The \(\beta_{best}\) values of the combined calibrated and random model predictor that maximize the ROC-AUC and PR-AUC respectively are \(\beta_{best}^{\text{ROC-AUC}}=-1.1\) and \(\beta_{best}^{\text{PR-AUC}}=-1.3\). For \(\beta=-1\) we still see significant performance improvement.
Best ROC and PRC
For both the Bliss and HSA ensemble-wise results we demonstrated above that a value of \(\beta_{best}=-1\) can result in significant performance gain of the combined predictor (\(calibrated + \beta \times random\)). So, the best ROC and PR curves we can get with our simulations when using models with both link operator (balance) and topology mutations are:
best_beta = -1
pred_topolink_ew_hsa = pred_topolink_ew_hsa %>% mutate(best_score = ss_score_150sim + best_beta * prolif_score_150sim)
pred_topolink_ew_bliss = pred_topolink_ew_bliss %>% mutate(best_score = ss_score_150sim + best_beta * prolif_score_150sim)
roc_best_res_hsa = get_roc_stats(df = pred_topolink_ew_hsa, pred_col = "best_score", label_col = "observed")
roc_best_res_bliss = get_roc_stats(df = pred_topolink_ew_bliss, pred_col = "best_score", label_col = "observed")
pr_best_res_hsa = pr.curve(scores.class0 = pred_topolink_ew_hsa %>% pull(best_score) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_hsa %>% pull(observed), curve = TRUE, rand.compute = TRUE)
pr_best_res_bliss = pr.curve(scores.class0 = pred_topolink_ew_bliss %>% pull(best_score) %>% (function(x) {-x}),
weights.class0 = pred_topolink_ew_bliss %>% pull(observed), curve = TRUE)
# Plot best ROCs
plot(x = roc_best_res_hsa$roc_stats$FPR, y = roc_best_res_hsa$roc_stats$TPR,
type = 'l', lwd = 3, col = my_palette[1], main = TeX('ROC curve (Ensemble-wise), $calibrated + \\beta \\times random$'),
xlab = 'False Positive Rate (FPR)', ylab = 'True Positive Rate (TPR)')
lines(x = roc_best_res_bliss$roc_stats$FPR, y = roc_best_res_bliss$roc_stats$TPR,
lwd = 3, col = my_palette[2])
legend('bottomright', title = TeX('AUC ($\\beta$ = -1)'),
col = c(my_palette[1:2]), pch = 19, cex = 1.5,
legend = c(paste(round(roc_best_res_hsa$AUC, digits = 2), 'HSA (150 sim)'),
paste(round(roc_best_res_bliss$AUC, digits = 2), 'Bliss (150 sim)')))
grid(lwd = 0.5)
abline(a = 0, b = 1, col = 'lightgrey', lty = 'dotdash', lwd = 1.2)
# Plot best PRCs
plot(pr_best_res_hsa, main = TeX('PR curve (Ensemble-wise), $calibrated + \\beta \\times random$'),
auc.main = FALSE, color = my_palette[1], rand.plot = TRUE)
plot(pr_best_res_bliss, add = TRUE, color = my_palette[2])
legend('topright', title = TeX('AUC ($\\beta$ = -1)'), col = c(my_palette[1:2]), pch = 19, cex = 1.5,
legend = c(paste(round(pr_best_res_hsa$auc.davis.goadrich, digits = 2), 'HSA (150 sim)'),
paste(round(pr_best_res_bliss$auc.davis.goadrich, digits = 2), 'Bliss (150 sim)')))
grid(lwd = 0.5)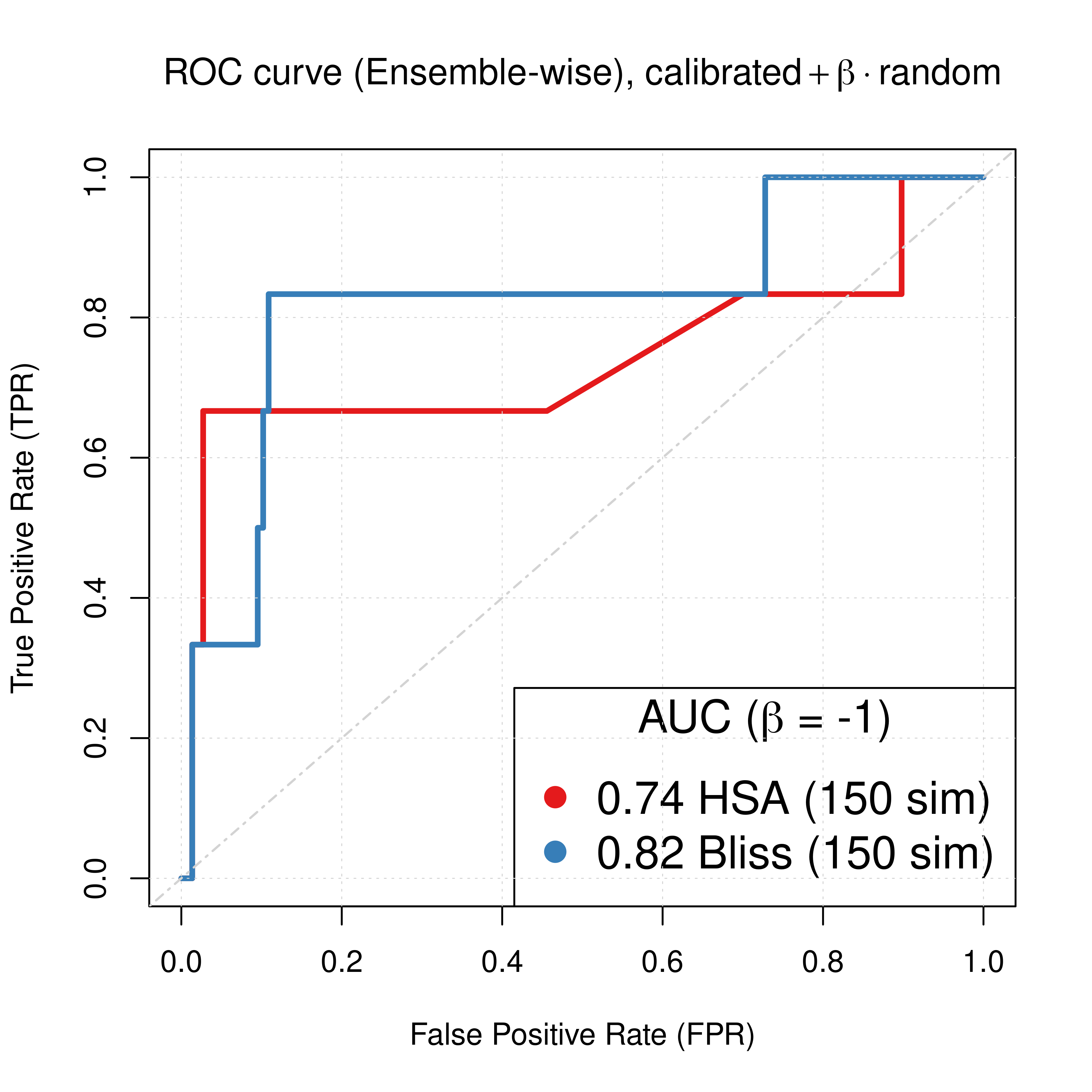
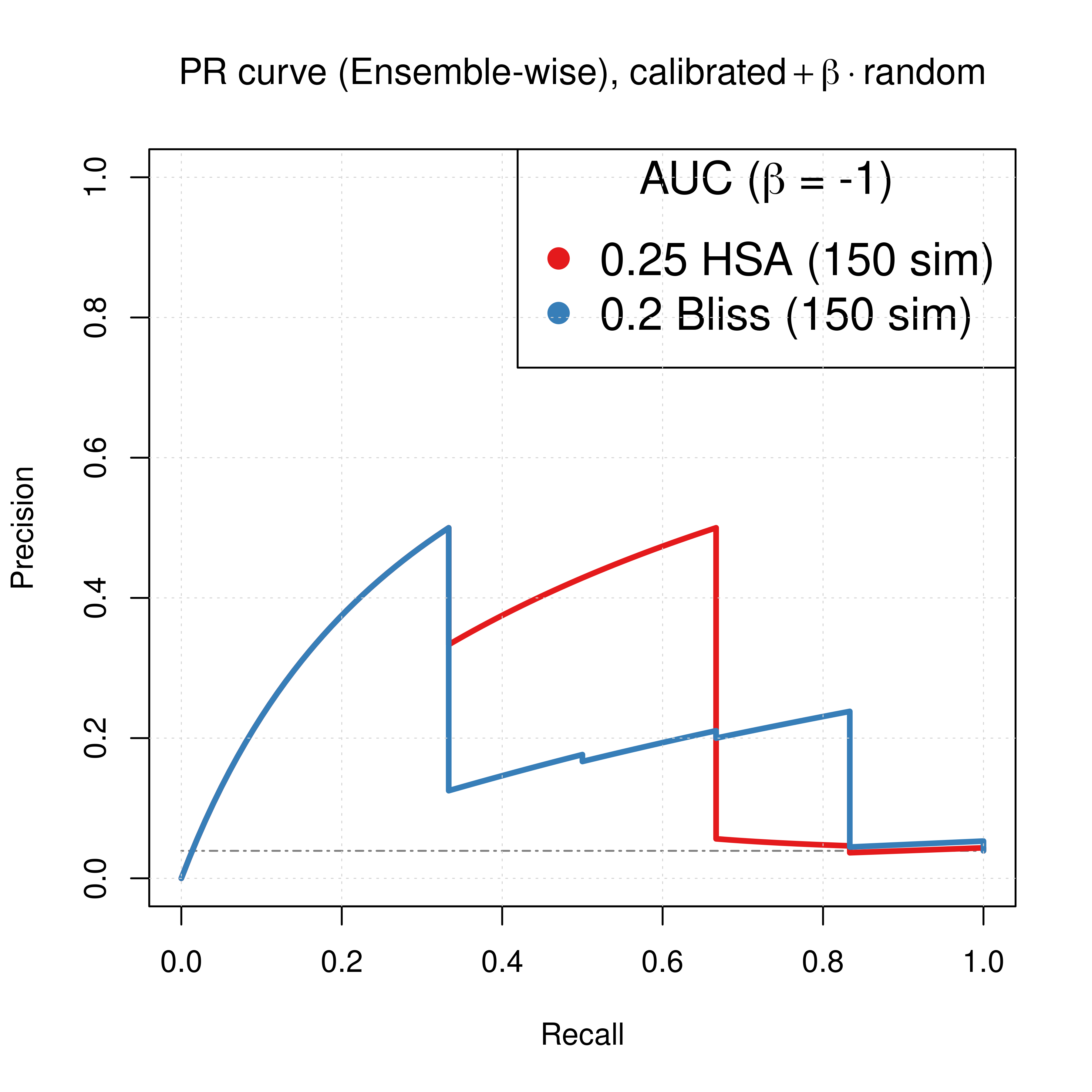
Figure 64: ROC and PR curve for best beta (CASCADE 2.0, Link Operator and Topology Mutations)